HiBiT-Powered CRISPR Knock-Ins for Endogenous Tagging
One Tiny Tag, Big Impact
CRISPR/Cas9 knock-in technology coupled with the tiny, powerful HiBiT tag allows researchers to study proteins in their natural environment. By eliminating the need for overexpression or antibody-based detection, HiBiT knock-ins preserve natural regulation and provide data that is more accurate, reproducible, and biologically meaningful using simple, scalable bioluminescent-based detection. Researchers can precisely quantify protein abundance using endpoint or live-cell kinetic methods, creating a clearer view of how proteins respond under changing conditions or therapeutic compound treatment. HiBiT technology is also compatible with surface-expressed and secreted protein detection. In addition, a sensitive, high-affinity anti-HiBiT monoclonal antibody enables further use in traditional immunoassays like Western blots and immunoprecipitation, providing additional ways to analyze HiBiT knock-in cells.
Our CRISPR/Cas9 knock-in cell lines support a wide range of applications in drug discovery and functional biology, including:
- Receptor internalization: track movement of membrane proteins and their regulation in live cells.
- Downstream signaling: measure how protein activity influences broader pathways.
- Targeted protein degradation: monitor degrader activity, kinetics and compound potency in real time.

Getting Started with CRISPR/Cas9 Knock-In Cell Lines
Choosing a workflow for CRISPR-enabled endogenous biology experiments will depend on the type of experiment, laboratory resources and experience, and the project timeline. Get started quickly by taking advantage of our collection of 250+ CRISPR-knock-in cell line pools and clones, available for a variety of target proteins and cell backgrounds. If you need to create custom knock-in cells, you have options. Check out the detailed protocol and access the HiBiT sequence. Need guidance? Reach out to one of our experts to ensure you get the results you need. Or, connect with a licensed service provider to explore custom knock-in cell generation options.
Choose the Path that Best Meets Your Needs

Check out our comprehensive list of 250+ cell line pools and clones for popular targets and cell backgrounds.


Use our detailed protocol to create your own HiBiT knock-in cells. Obtain the HiBiT sequence and synthesis rights and review our terms of use by completing our licensing agreement.
Collaborate with experts who offer custom knock-in and gene-editing services:
EditCo Bio – Provides functionally validated CRISPR-edited cell lines, offering precise and scalable genome engineering solutions to support research in cell and gene therapy, drug discovery, and basic biology.
FUJIFILM CDI – Offers custom knock-in cell line generation using human iPSC-derived cells to create physiologically relevant models for disease research, drug discovery, and cell therapy development.
ALSTEM, Inc. – Delivers custom iPSCs, CRISPR/Cas9-edited cell lines, and viral vectors, supporting research in regenerative medicine, disease modeling, drug discovery, and basic biology.
Have a question?
Why Choose HiBiT for CRISPR Knock-In Experiments?
HiBiT knock-in tagging combines the precision of CRISPR with the sensitivity of bioluminescence. By inserting HiBiT directly into the genome, you can:
- Monitor physiological expression to avoid gene dosage effects and artifacts from overexpression.
- Study genes under natural promoter and epigenetic control.
- Maintain protein balance by preserving natural ratios with interacting partners.
- Generate knock-in models faster as no molecular cloning is required.


Hear from a Scientist
Discover how researchers like Dan Nomura, Ph.D. from University of California - Berkeley are using HiBiT-powered CRISPR knock-ins to study protein dynamics at endogenous levels.
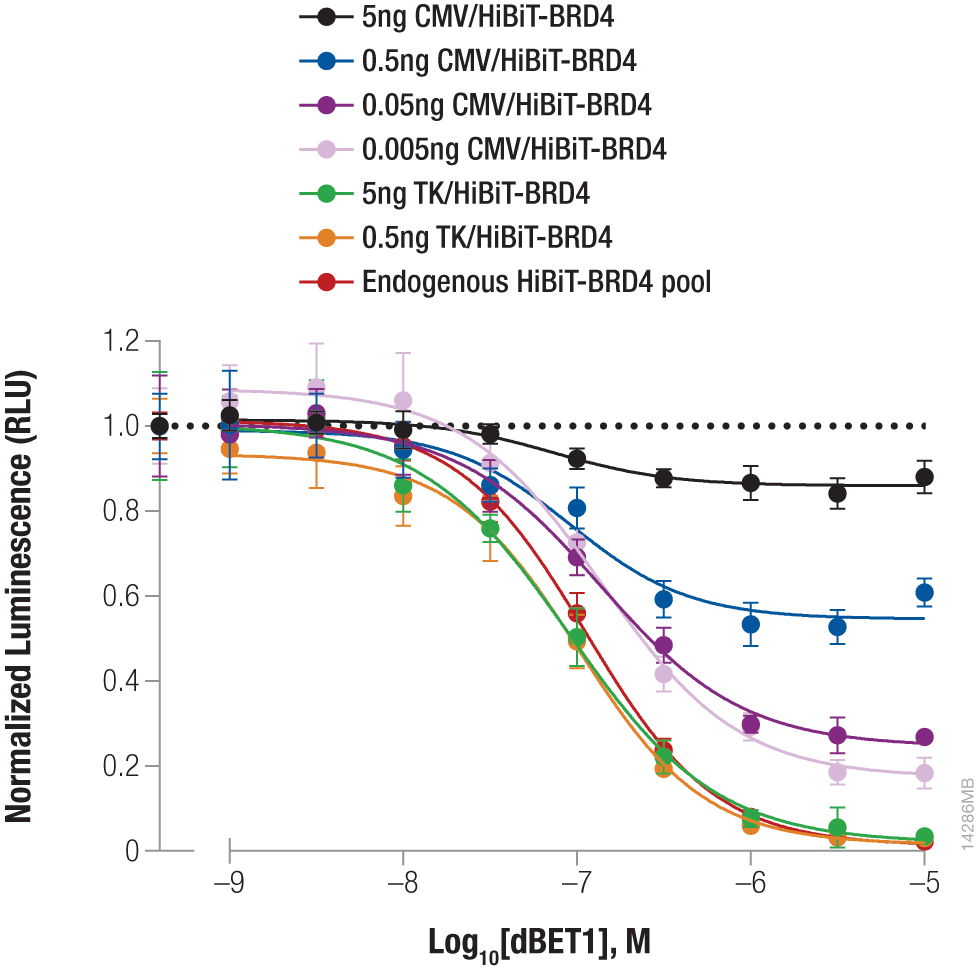
Endogenous tagging creates an improved assay window for BRD4 degradation assay
Protein levels shift with cellular conditions or therapeutic interventions, such as protein degrader treatments, making sensitive assays essential. Here, we used a CRISPR knock-in of HiBiT at the BRD4 locus to measure protein abundance following treatment with the dBET1 degrader. BRD4 under native regulation was compared with different dilutions of CMV or TK-driven overexpression. Cells were treated with a titration of dBET1 and protein abundance was measured by quantifying the HiBiT tag (using the Nano-Glo® HiBiT Lytic Detection System). Normalized luminescence values demonstrated that the knock-in cells provided an increased assay window when compared to higher level overexpression models, highlighting the advantage of monitoring native expression.
Applications of CRISPR Knock-In Gene Editing
Targeted Protein Degradation

Selectively targeting proteins for removal from cells is a promising new modality for potential therapy. Watch this webinar to learn about pairing CRISPR-knock-in cell lines with TPD studies.
Gene Expression Modulation

Oligotherapeutics have advanced rapidly, but highthroughput, quantitative gene expression measurement remains a challenge. View our poster explaining more about these solutions.
Protein Dynamics

Studying protein dynamics is often slow and disruptive. Here, we show how a single HiBiT knock-in can track localization, interactions, and kinetics of a key chromatin remodeler. Read the article to learn more.
Resources for HiBiT-Tagging in CRISPR Cell Lines
Webinar
Understanding Your Options with CRISPR-Mediated Tagging.
Article
Learn How Researchers Tag Endogenous Proteins Small, Luminescent Peptides.
Application Note
Create Reporter Knock-In Cells with Our Optimized Guide.
FAQ
Answers to common questions about HiBiT tagging, CRISPR knock-ins, assay setup and more.